Segmentation¶
We provide different plugins for different segmentation tasks:
- Install plugin
SegmentObjectsto segment cells OR nuclei from here. - Install plugin
SegmentCellsNucleito segment cells AND nuclei from here.
General behavior¶
Recursive search¶
The plugins will search recursively the provided subfolder for images to segment.
By default, all images matching the naming scheme will be processed. An optional parameter allows to specify in which subfolder the images have to be in order to be segmented. This allows to process nested folder hierachies and only segmenting images in the relevant subfolders.
As an example, in the provided data we could specif the folder example_data\analysis as the data folder.
This would then scan this folder and all subfolders and potentially find inappropriate files for segmentation.
By defining the Input subfolder to be segmentation-input, the analysis will be restricted to this folder.
Results¶
Results will be saved in the specified folder. For each image the following files, results files with different suffixes are created:
..._flow: these are the predicted distance maps of CellPose. They are an intermediate result, and not needed for most end-users...._mask: these contain the actual segmentation results. Each segmented object is a filled object with a constant pixel value. If the images were resized during segmentation, the mask is scaled back up to the original image size. The actually obtained (smaller) mask is saved under the namemask__rescale_......._seg: summary plot showing the input image, the predicted distance map, and the segmented objects. This plot is also shown in the interface.
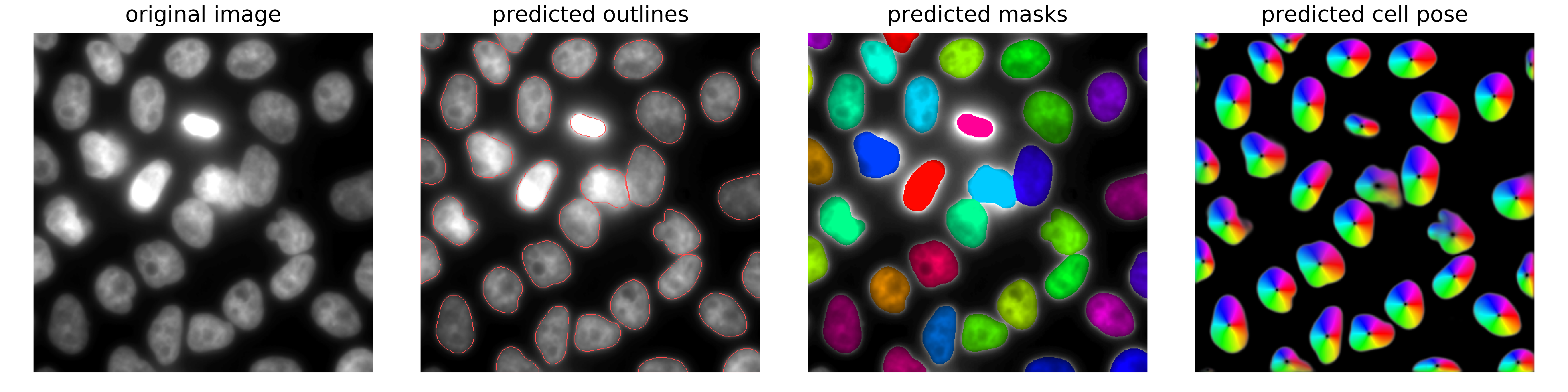
Resizing can speed up prediction & yield better results¶
We found that resizing images before segmentation can yield better results for certain images. Also, it will lead to faster computation time. In case you resize the images, we implemented a post-processing routine that will resize the predicted masks back to the original image size.
Resizing can be specified in two ways
- Scaling factor recommended]: simply add an integer value. The actual image size will then be divided by this factor to obtain the new size, i.e. a value of 2 will resize an image 512x512 to 256x256.
- New size: you can directly define the new size of the image, e.g. 256x256. Please note that this size will be applied to all images, independly of their size. This option is hence not suitable if your data-sets contain differently sized images.
Recommended workflow¶
The default settings of the plugins allow to quickly perform the recommended workflow. You only have to paste your data folder.
- 2D images are stored in a subfolder
segmentation-input - Segmentation results will be stored in a subfolder
segmentation-results, this can be achieved by setting the save path to the string replacementsegmentation-input>>segmentation-results
Segmentation of nuclei OR cells¶
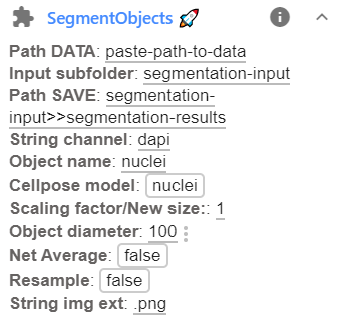
-
Before running the plugin, you have to specify a few parameters. This can be done in the plugin interface, avaible after clicking on the arrow down next to the plugin name.
Here the following parameters can be set:
Option Type Default Description Path DATAstr Full path to folder containing data to be segmented. Input subfolderstr Name of the subfolder containing the images that should be segmented. Path SAVEstr Several options exist. See dedicated section below for more details. String channelstr dapiUnique identifier to identify channel. Object namestr nucleiHow the object is called. Cellpose modelstr nucleiCellpose model for segmentation: cytoornuclei. Note that for dense nuclei, the cytoplasmic model might work better.New sizeint 2 Numbers to specify resizing (see above). No resizing if empty. Object diameterint 50 Typical diameter of the object. Better to be set a bit to small. Net AverageBool False Can improve segmentation accuracy, but is slower (Runs the 4 built-in networks and averages them). ResampleBool False Gives more accurate boundaries, but can be very slow (Runs dynamics at original image size). String img extstr .pngFile extension of images that should be segmented. New sizeint 2 Numbers to specify resizing (see abov. No resizing if empty. -
Pressing on the plugin name
SegmentObjectswill start the segmentation. When using CellPose for the first time, the models for nuclear and cytoplasmic segmentations are downloaded.The actual segmentation can take a while, depending on the numberof images that should be segmented (and their size). Progress will be displayed in the ImJoy status bar, and more details provided in the plugin log available by pressing on the
inext to the plugin name.Once a image is segmented, the results will be saved (see below). So you can monitor the result folder to verify on the fly if the segmentation works.
Segmentation of cells AND nuclei¶
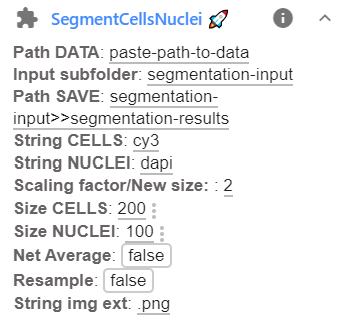
-
Before running the plugin, you have to specify a few parameters. This can be done in the plugin interface, avaible after clicking on the arrow down next to the plugin name.
Here the following parameters can be set:
Option Type Default Description Path DATAstr Full path to folder containing data to be segmented. Input subfolderstr Name of the subfolder containing the images that should be segmented. Path SAVEstr Path to folder where results should be stored (for more details see above). String CELLSstr cy3Unique identifier for images of cytoplasmic stain. String NUCLEIstr dapiUnique identifier for images of nuclear stain. String img extstr .pngFile extension of images that should be segmented. New sizeint 2 Numbers to specify resizing (see above). No resizing if empty. Size CELLSint 100 Typical size of a cell (in resized image). Size NUCLEIint 50 Typical size of a nucleus (in resized image). Net AverageBool False Can improve segmentation accuracy, but is slower (Runs the 4 built-in networks and averages them). ResampleBool False Gives more accurate boundaries, but can be very slow (Runs dynamics at original image size). String img extstr .pngFile extension of images that should be segmented. -
Pressing on the plugin name
SegmentCellsNucleiwill start the segmentation. When using CellPose for the first time, the models for nuclear and cytoplasmic segmentations are downloaded.The actual segmentation can take a while, depending on the numberof images that should be segmented (and their size). Progress will be displayed in the ImJoy status bar, and more details provided in the plugin log available by pressing on the
inext to the plugin name.Once a image is segmented, the results will be saved (see below). So you can monitor the result folder to verify on the fly if the segmentation works.